
Genomic information is important for a range of areas in public health protection, from tracking infectious disease outbreaks, to identifying inherited disorders, and characterising mutations that underpin the progression of cancers.
Genome: An organism’s complete set of DNA, including all of its genes
Microorgansim/microbe: A microscropic living organism including bacteria, fungi and viruses
Isolate: A culture of microorganisms isolated for study
What are the applications of WGS for microorganisms?
One of the most important roles for genome sequencing is when it is applied to microorganisms, also referred to as pathogens. In this realm it has multiple applications, with the most notable being infectious disease management at both individual and population level.
DNA profiling has traditionally been used to determine the likelihood that genetic material has come from a particular individual or group. In addition to this, WGS also enables:
- rapid identification and characterisation of microorganisms, providing information on the relatedness of strains, where they have come from and how they have evolved
- identification of key virulence factors – specific chacteristics that aid the microorganism in causing infection
- antibiotic resistance profiling – compared with traditional culture methods, WGS can determine which antibiotics microorganisms are resistant to much more quickly
- detection, mapping and analysis of outbreaks
As a highly discriminatory and accurate technique that is equivalent to forensic-level typing, WGS provides the most robust microbioogical evidence. Scientists at PHE focus on sequencing the genomes of microorgansims, however it is important to note that the technology also has important applications for humans, such as:
- developing personalised medicine and tailoring medical treatment so that it fits the specific characteristics of the patient
- characterising cancer tumours to identify the most effective treatment for halting tumour growth with minimal toxicity to the patient
- diagnosing rare diseases and identifying the causative genetic variant that is responsible for them, leading to further studies of gene targets to characterise disease mechanisms
Surveilling infection outbreaks
WGS has become an integral part of the surveillance of infection disease, including foodborne illness outbreaks.
The genome sequences of bacteria from infected individuals are compared to identify clusters of related organisms that have highly similar genomes. Once a cluster is detected, the next step involves investigating whether they originated from the same source through finding out about the recent history of infected individuals. The full process is outlined further down.
Analysing microorganisms’ susceptibility to antibiotics
WGS can be used to characterise specific features encoded in a microorganism’s genome. This can be used to predict the bacterium’s resistance to antibiotics and can also shed light on specific features that aid the microorganism in causing infection.
As a result, treatment regimens can be tailored accordingly, such as treatments for tuberculosis (TB) when the causative pathogen may be resistant to the first line drugs. Tailoring the patient’s treatment will significantly improve their chances of a full recovery and help reduce the spread of drug resistant bacteria.
The different methodologies
WGS is the umbrella term for DNA sequencing, but there are different methodologies that can be used to sequence the genome, including:
- Short read sequencing, or sequencing by synthesis (SBS)
- Single molecule real time sequencing (SMRT)
Short read sequencing is commonly provided by Illumina. It is the predominantly used method responsible for generating more than 90% of the world’s sequencing data, and currently the most effective method for looking at individual differences between genomes. It provides short read data by fragmenting the DNA into small sections.
On the contrary, SMRT does not fragment the DNA prior to sequencing, thus providing very long read data. Although this method is currently less accurate than SBS, some models such as the Oxford Nanopore are small and highly portable, therefore permitting sequencing in the field.
WGS in the field
PHE scientists took a Nanopore sequencer to Sierra Leone, Liberia and Guinea during the Ebola outbreak to sequence new Ebola cases. Sequencing in the field is faster than shipping specimens back to laboratory in another country, and this is ideal because timely interventions have more impact on public health.
The multidisciplinary process of WGS at PHE
Here at PHE, we have a multidisciplinary team of scientists who carry out the multiple steps of WGS:
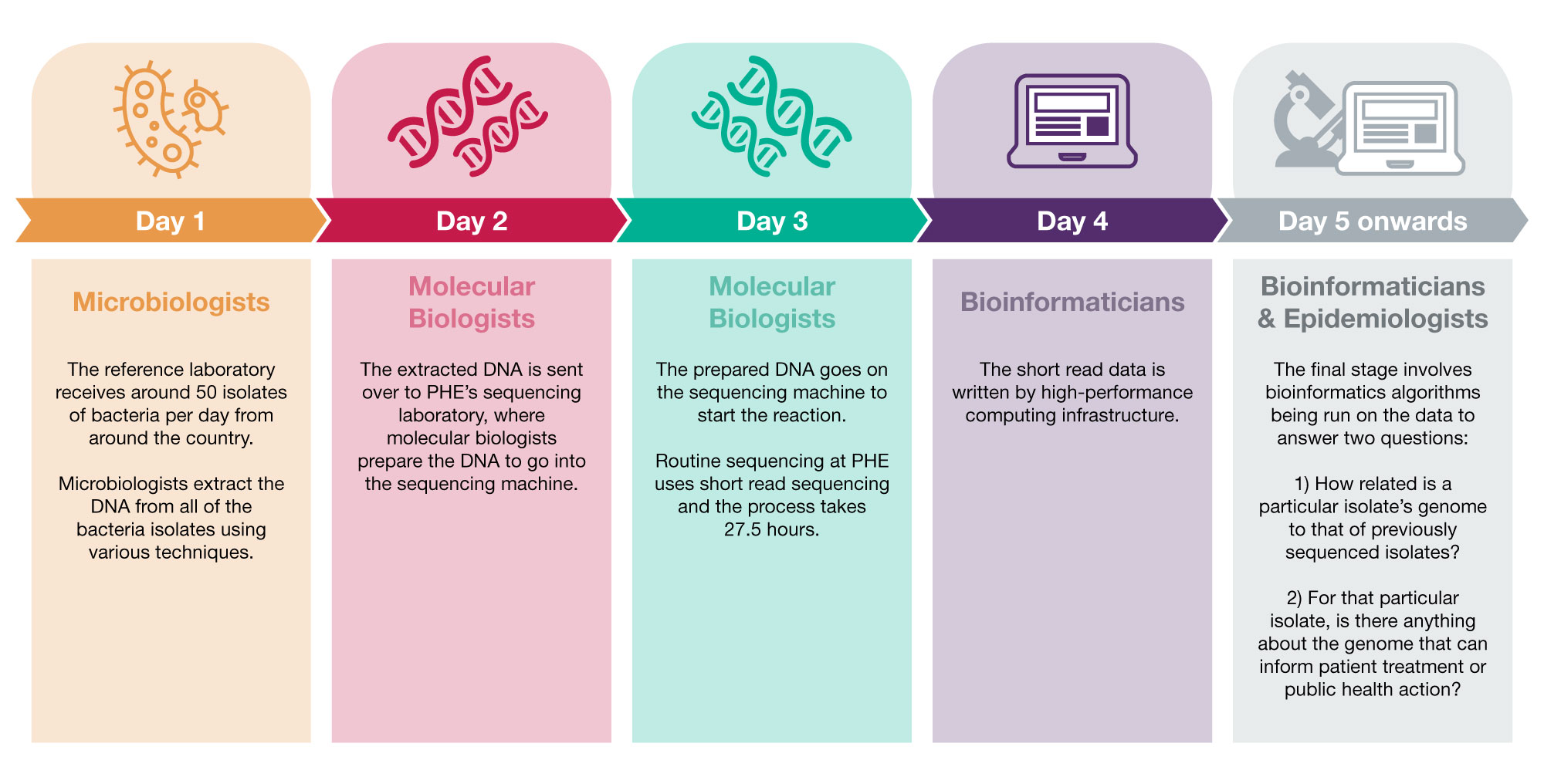
Once we have sequenced a new set of bacterial genomes we compare them to a database of all the previously sequenced bacterial genomes from routine pathogen surveillance at PHE. As no two bacteria have the same genome by chance, if there is a match the patients from which the bacteria were isolated from may share a common exposure. For a foodborne pathogen like Salmonella it may mean they have eaten the same food or attended the same restaurant.
Epidemiologists use surveillance questionnaires to gather information from the people that the isolates came from, to build up a picture of anything they have in common. This is particularly important for identifying the source responsible for complex, nationally distributed outbreaks, such as a product sold in a major supermarket. With the globalisation of the food chain, this forensic-level analysis is crucial.
The future of WGS
WGS will continue to become technically and economically feasible as time goes on, and will therefore have an increasingly positive impact on the practice of medicine and public health. Pathogen sequencing, in particular, will have an progressively important role in health protection practice as it reduces and manages disease outbreaks more efficiently and effectively allowing better public health control and prevention actions to be put into place.
PHE is already pioneering the development and early use of WGS for the investigation and control of multiple infectious diseases. WGS will continue to be a priority area of focus for PHE in the future, and we will work to be at the forefront of this technology.
Identifying different strains of disease-related microorganisms
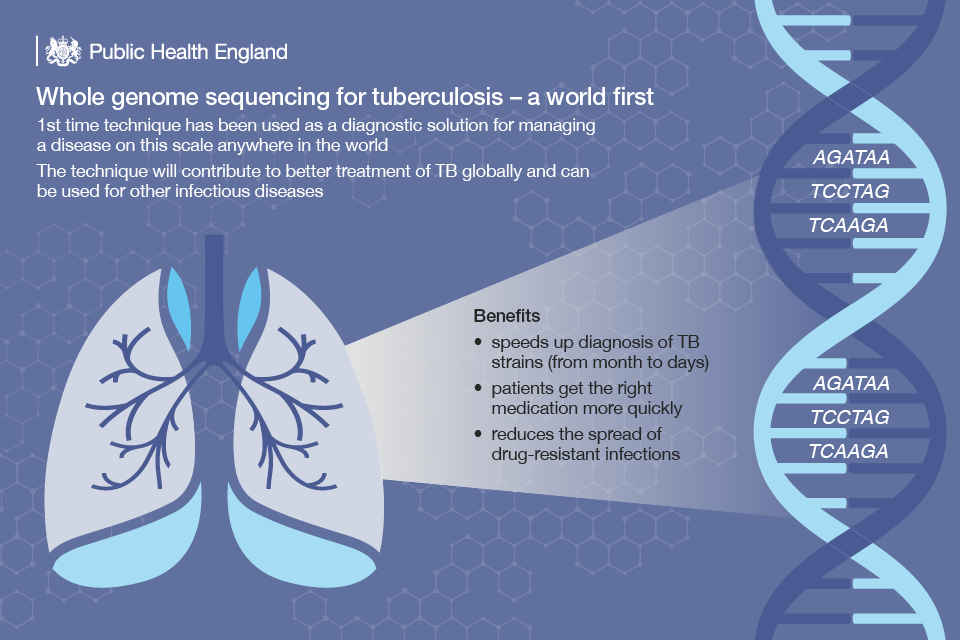
At the end of 2016, we began using WGS to identify different strains of TB, the first time that this has been used to diagnose and manage a disease at this scale anywhere in the world.
Where previously it could take up to a month to confirm a diagnosis of TB and the treatment choices, and to detect spread between cases, this can now be done in just over a week at PHE’s Birmingham laboratory. This slows the spread of the disease and boosts the fight against antimicrobial resistance.
WGS is being used to identify strains of TB that are drug resistant, which need to be treated differently to a non-resistant infection. In less than 24 hours, we can find out if a specific infection is likely to be resistant to a particular drug and recommend a different course of treatment.
Sequencing in the field and by the bedside
In future, the application of portable sequencing for infectious diseases and conditions may allow us to test and treat patients at the bedside, rather than needing to wait days and sometimes weeks for results and sending samples between locations.
The implementation of WGS will contribute to achieving the government’s ambition of sequencing 100,000 genomes, a project which will ultimately allow us to better understand major diseases and how to treat them.
